Document Type : Original Article
Authors
1 Department of Food Hygiene and Aquaculture, Faculty of Veterinary Medicine, Ferdowsi University of Mashhad, Mashhad, Iran
2 Antimicrobial Resistance Research Center, Avicenna Research Institute, Tehran, Iran
3 Department of Microbiology and Virology, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
4 Department of Pathobiology, School of Medicine, North Khorasan University of Medical Sciences, Boujnord, Iran
Abstract
Staphylococcus aureus is generally regarded as a leading cause of mastitis in dairy cattle. The aim of this study was to investigate the pattern of agr groups and any possible relationship between agr groups and antibiotic resistance among S. aureus strains isolated from bovine mastitis in Northeast of Iran. For this purpose, a total of 300 bovine mastitic milk samples were taken from dairy industry farms of Khorasan Razavi Province, Iran. S. aureus were isolated and identified according to the standard methods. Antibiotic susceptibility testing was conducted by disk diffusion method. In this study a total of 31 isolates of S. aureus were evaluated for agrD gene polymorphism by specific primers. Most of the isolates belonged to agr group I (54.8%), followed by agr group III (25.8%) and agr group II (19.4%). There was not any isolates belonging to group IV. Resistance to methicillin in agr group I isolates was more than other groups. Agr groups II and III were quite susceptible to methicillin. Due to high prevalent of S. aureus isolates and high antibiotic resistance rate in bovine mastitic isolates, it is important to verify the characteristics of S. aureus strains in Iran.
Keywords
Main Subjects
Introduction
Staphylococcus aureus is a common cause of subclinical mastitis worldwide, which is of economic importance to the dairy industry.1 The main reservoir of S. aureus in milk and milk products seems to be infected quarter. Molecular epidemiological analysis of the bovine S. aureus population suggested that a small number of clonal types were responsible for most infections, and that isolates had a broad geographic distribution.2-4 Staphylococcus aureus has a capacity to produce a large number of putative virulence factors. Some of these factors may be more important than others in different diseases or at different stages of the pathogenesis of particular infections, as not all factors are produced by each strain.5-7
Recently the accessory gene regulator (agr) locus was identified as a regulator of virulence factorsin S. aureus. It controls a large set of genes including those encoding cell wall associated and extracellular proteins.8
The agr locus is composed of two divergent transcriptional units, RNAII and RNAIII driven by the P2 and P3 promoters respectively. The P2 operon encodes four proteins (agrA, agrB, agrC and agrD) that generate the agr-sensing mechanisms. The agr signaling system consists of a classical two-component regulatory system in which the agrC (the signal receptor) bind the extracellular auto inducing peptide, agrD (AIP) and modulates the activity of agrA, the response regulator. agrA activity then leads to greatly increased P2 and P3 transcription. agrB is a transmembrane protein that involved in processing and secreting of the agrD. Sequence diversity in the variable region, comprised of the last one-third of agrB, agrD and the first half of agrC, has generated the four agr specificity group in S. aureus. The AIP produced by a given strain of S. aureus activates its own agr locus but may inhibit the expression of agr in other isolates. This AIP-dependent inhibition is correlated with ability of a strain to compete with other isolates for sites of infection. RNAIII encodes several virulence factors, including TSS toxin1 and hemolysins. The Staphylococcal agr system also decreases the expression of several cell wall-associated proteins.8-10
In this research we study the pattern of agr groups among S. aureus isolates obtained from bovine mastitis and investigate any possible relationship between agr groups and antibiotic resistance in this region.
Materials and Methods
Bacterial isolates. Milk samples (10 mL) were taken aseptically from all quarters of 300 bovine infected udders of some dairy industry farms of Khorasan Razavi, Northeast of Iran. The presence of Staphylococcus spp. was determined by culturing 0.01 mL of each sample on 5% bovine blood agar (Merck Millipore, Darmstadt, Germany) plates and incubated at 37 ˚C for 24 to 48 hr. A quarter was identified as infected when a single pathogenic bacterium was isolated and somatic cell count (SCC) was increased above 200,000 mL-1.
The bacterial isolates were presumptively identified on the basis of morphology, catalase production, hemolysis pattern and Gram staining of the colonies, coagulase activity on rabbit plasma (Bio-Merieux, Marcy l'Etoile, France), mannitol fermentation on mannitol salt agar (MSA; Himedia Labs, Mumbai, India), production of clamping factor (Slidex Staph Plus; Bio-Merieux, Marcy l'Etoile, France) and each bacterial isolates was streaked on blood agar to obtain a pure culture.
Antimicrobial susceptibility. Antibiotic susceptibility testing was conducted by disk diffusion method according to the guidelines of the National Committee for Clinical Laboratory Standards.11
DNA extraction and amplification. The genomic DNA of S. aureus isolates was extracted by the modified phenol-chloroform method. Lysates of colonies were prepared according to protocol given by Sharma et al. 12
The specific agr groups were determined by poly-merase chain reaction (PCR) with use of pan forward agr: 5'-ATGCACATGGTGCACATGC-3' corresponding to conserved region from agrB gene, Reverse agr I: 5'-GTCACAAGTACT ATAAGCTGCGAT-3' (in the agrD gene, product size 440 bp), Reverse agr II: 5'-GTATTACTAATTGAAAAGTGCCATA GC-3' (in the agrC gene, product size 572 bp), Reverse agr III: 5'-CTGTTGAAAAAGTCAACTAAAAGC TC-3' (in the agrD gene, product size 406 bp) and Reverse agrIV: 5'-CGATAA TGCCGTAATACCCG-3' (in the agrC gene, product size 588 bp).13 The PCR assay was performed by adding 1 µL of a 1:200 dilution of S. aureus DNA template and 24 µL of water to 25 µL of a PCR mixture that includes 2.5 IU of Taq DNA polymerase, 2 mM MgCl2, 350 µm dNTPs and 25 mM KCl in 0.2 mL PCR tubes. Thermal cycling was performed in a thermal cycler (Techne, Cambridge, UK) and consisted of 30 cycles of denaturation (94 ˚C in 60 sec), annealing (57 ˚C in 60 sec) and extension (72 ˚C 60 sec). Aliquots of amplified samples were analyzed by electrophoresis on a 1.0% agarose gel and stained with ethidium bromide. S. aureus strains RN6390 (agr group I), RN6607 (agr group II), RN8465 (agr group III), RN4550 (agr group IV) and RN6911 (agr negative) were used as positive control for agr group identification, kindly provided by Dr. Richard P. Novick (Skirball Institute of Biomolecular Medicine, New York, USA).
Statistical analysis. Statistical differences between groups were analyzed by means of student’s t-test or analysis of variance (ANOVA) tests in SPSS (version 17; SPSS Inc., Chicago, USA). Multivariate analysis was performed to assess the independence of the statistically significant variables in unvaraiate analysis. A p-value less than 0.05 was considered significant.
Results
In this study a total of 31 isolates from 300 samples were evaluated. Analysis of agrD gene polymorphism by specific primers allowed assigning our isolates in one of four major specific agr groups. The agr specific groups were determined by mentioned PCR method (Fig. 1).
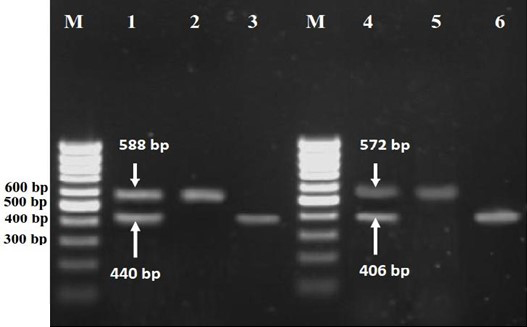
Fig. 1. Agarose gel electrophoresis of PCR products for agr specific groups. Lane M, 100 bp DNA molecular size marker; Lane 1, Control positive (combined PCR products of RN6390 (440 bp for agrI group,) and RN4550 (588 bp for agrIV,group) reference strains); Lane 2, PCR product of agrII group; Lane 3, PCR product of agrI group; Lane 4, Control positive (combined PCR products of RN6607 (572 bp for agrII group,) and RN8465 (406 bp for agrIII,group) reference strains); Lane 5, PCR product of agrII group and lane 6, PCR product of agrIII group.
Most of our isolates belonged to agr group I (54.8%), followed by agr group III (25.8%) and agr group II (19.4%). There was not any strain belonging to group IV.
Considering the possible relation between the agr group and the antibiotic resistance in S. aureus isolates, we used disk diffusion agar test according to National Committee for Clinical Laboratory Standards (NCCLS) and the results were demonstrated in Table 1.
According to these results, there is no significant association between resistance against penicillin and agr group but there is a significant association between agr group and resistance against methicillin.
Table 1. Frequency of agr groups and their antibiotic resistance among Staphylococcus aureus isolates. The values indicate the number (frequency) of isolates.
|
Antibiotic |
agr groups |
Total (n = 31) |
||
|
agr group I (n = 17) |
agr group II (n = 6) |
agr group III (n = 8) |
||
|
Penicillin |
15(88.2%) |
5(83.3%) |
8(100%) |
28(90.3%) |
|
Ampicillin |
12(70.6%) |
3(50.0%) |
6(75.0%) |
21(67.7%) |
|
Cefalotin |
1(5.9%) |
1(16.7%) |
1(12.5%) |
3(9.7%) |
|
Cefixime |
8(47.0%) |
2(33.3%) |
2(25.0%) |
12(38.7%) |
|
Tetracycline |
5(29.4%) |
0 |
1(12.5%) |
6(19.4%) |
|
Gentamycin |
0 |
0 |
0 |
0 |
|
Methicillin |
9(52.9%) |
0 |
0 |
9(29.0%) |
|
Vancomycin |
0 |
0 |
0 |
0 |
Discussion
Staphylococcus aureus is a major pathogen in both human and animals.14 The ability of this organism to cause multitude of human disease such as endocarditis, pneumonia, bacteremia and toxic shock syndrome (TSS) and diseases of farm animals such as mastitis suggested that the pathogenesis of Staphylococcus aureus infections is highly complex. The virulence of organism is dependent on many cell surface proteins and secreted exotoxins and enzymes and it is suggested that the environmental and host signals are contributing the regulation of virulence factors.15 The agr operon is involved in the coordinate regulation of some of Staphylococcus aureus virulence factors. Staphylococcus aureus isolates exhibit well-defined genetic polymorphisms within the agr locus. Four agr genotypes, group I to IV have been described to date.16
Takeuchi et al. have shown that among 76 S. aureus isolated from cow mastitis in Japan, 43.4% isolates belonged to agr group I , 18.4% belonged to agr group II and 38.2% belonged to agr group III.17 This report is in consistent with our findings which showed agr group I (54.8%), agr group II (19.4%) and agr group III (25.8%).
Gilot et al. reported that among 71 S. aureus isolated from bovine mastitis 69.0% of isolates belonged to agr group I , 23.9% belonged to agr group II, 2.8% were agr group III and 1.4% were belonged to agr group IV that were somewhat different from our results and more similar to the human isolates.18
Although data relating agr type and specific infections are scarce, Jarraud et al. have shown that specific agr genotype isolates might be associated with particular infectious syndromes.15 For example, disease mediated by enterotoxin is linked to agr group I, infective endocarditis is linked to agr groups I and II, toxic shock syndrome is linked to agr group III and exofoliative disease is linked to agr group IV. Recently, agr group III genotype isolates have been over represented in isolates from community-acquired methicillin-resistant S. aureus (MRSA) infections, whereas previous nosocomially isolated MRSA isolates in the United States were predominantly of agr group II.19,20 Most exofoliatin producing isolates responsible for staphylococcal scalded skin syndrome (SSSS) belonged to agr group IV.21
In our study resistance to methicillin in agr group I isolates was more than other groups. Agr groups II and III were quite susceptible to methicillin. Furthermore, other studies showed a correlation between induction of the glycopeptide intermediate S. aureus (GISA) phenotype and autolytic deficiency, especially in the context of the agr genotype II,22 but in our study all isolates were susceptible to vancomycin. Some reports state that there are clinical trends according to each agr group. For example, agr group I was prevalent in a collection of 192 S. aureus isolates, 71.0% of which were methicillin resistant.16,21
Moise-broder et al. showed that agr group II poly-morphism in MRSA predicts the failure of vancomycin therapy.23 Reportedly, community-acquired MRSA belonged to agr group III and methicillin sensitive S. aureus (MSSA)24 and toxic shock syndrome toxin (TSST-1) producing isolates belonged to the agr specificity group III.17 Recent data demonstrate that the vast majority of MRSA in France and around the world belong to agr group III.24,25
In our study most of MRSA isolates were belonged to agr group I and all of MSSA isolates belonged to agr groups II and III. Agr specific group IV was absent in many previously reported.13,16,26 and also we do not have any group IV isolates in our isolates. This is more likely due to ecological and geographical structuring.
There seems to be a geographic difference between agr groups. Most isolates belonged to agr group I, represented by the Brazilian, Portuguese, Hungarian and Berlin. Group II isolates, represented by the pediatric and Japan isolates, have been isolated mainly in Japan and North America. Isolates of group III isolated mainly in Europe.27 Our isolates revealed that group I isolates are prevalent in Iran, followed by group III and group II, which was relatively small compared to the previous groups. Iran is one of several countries with high antibiotic resistance rate. Therefore it is important to verify the characteristics of S. aureus in this country. The results of this study showed the status of agr groups and antibiotic resistance patterns between S. aureus isolated from bovine mastitis in Northeast of Iran. This study may also aid finding an appropriate method to eradicate resistant clones because agr is a potential target for therapy and the response can be modulated by the synthetic peptides.
Acknowledgments
This research was supported by Research Council of the Ferdowsi University of Mashhad (Grant No. 33268). The authors acknowledge Dr. Richard P. Novick (Skirball Institute of Biomolecular Medicine, New York, USA) and Dr. Patrice Francois (University of Geneva Hospitals, Geneva, Switzerland) for supporting Standard strains and research suggestions.
- Kalorey DR, Shanmugam Y, Kurkure NV, et al. PCR-based detection of genes encoding virulence determinants in Staphylococcus aureus from bovine subclinical mastitis cases. J Vet Sci 2007; 8: 151-154.
- Fitzgerald JR, Meaney WJ, Hartigan PJ, et al. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol Infect 1997; 119: 261-269.
- Kapur V, Sischo WM, Greer RS, et al. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J Clin Microbiol 1995; 33: 376-380.
- Salasia SIO, Khusnan Z, Lammler C, et al. Comparative studies on pheno- and genotypic properties of Staphylococcus aureus isolated from bovine subclinical mastitis in central Java in Indonesia and Hesse in Germany. J Vet Sci 2004; 5: 103-109.
- Fitzgerald JR, Hartigan PJ, Meaney WJ, et al. Molecular population and virulence factor analysis of Staphylococcus aureus from bovine intramammary infection. J Appl Microbiol 2000; 88: 1028-1037.
- Foster SJ. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J Bacteriol 1995; 177: 5723-5725.
- Foster G, Ross HM, Huston RA, et al. Staphylococcus lutrae sp. nov., a new coagulase-positive species isolated from otters. Int J Syst Bacteriol 1997; 47: 724-726.
- Novick RP. Auto-induction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 2003; 48: 1429-1449.
- Wright JS, Traber KE, Corrigan R, et al. The agr radiation: an early event in the evolution of staphylococci. J Bacteriol 2005; 16: 5585-5594.
- Yarwood JM, Schielvert PM. Quorum sensing in Staphylococcus infections. J Clin Invest 2003; 11:
1620-1625. - National Committee for Clinical Laboratory Standards (NCCLS). Performance standards for antimicrobial disk susceptibility tests. 7th ed. Approved Standard M2-A7. Wayane, USA; 2000; 17-49.
- Sharma NK, Rees CED, Dodd CE. Development of a single-reaction multiplex PCR toxin typing assay for Staphylococcus aureus strains. Applied and Enviro Microbiol 2000; 66(4):1347-1353.
- Shopsin B, Mathema B, Alcabes P, et al. Prevalence of agr specificity groups among Staphylococcus aureus isolates colonizing children and their guardian's. J Clin Microbiol 2000; 1: 456-459.
- Bunce C, Wheeler L, Reed G, et al. Murine model of cutaneous infection with gram-positive cocci. Infect Immun 1992; 60: 2636-2640.
- Jarraud S, Mougel C, Thioulous J, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles) and human diseases. Infect Immun 2002; 70: 631-641.
- Manago K, Nishi J, Wakimoto N, et al. Biofilm formation by and accessory gene regulator typing of methicillin-resistant Staphylococcus aureus isolates recovered from patients with nosocomial infections. Infect Control Hosp Epidemiol 2006; 27: 188-190.
- Takeuchi S, Maeda T, Hashimoto N, et al. Variation of the agr locus in Staphylococcus aureus isolates from cows with mastitis. Vet Microbiol 2001; 79: 267-274.
- Gilot P, Van Leeuwen W. Comparative analysis of agr locus diversification and overall genetic variability among bovine and human Staphylococcus aureus isolates. J Clin Microbiol 2004; 42: 1265-1269.
- Naimi TS, LeDell KH, Como-Sabetti K, et al. Comparison of community and health care associated methicillin resistant Staphylococcus aureus infection. J Am Med Assoc 2003; 290: 2976-2984.
- Sakoulas G, Eliopoulos GM, Moellering RC, et al. Staphylococcus aureus accessory gene regulator (agr) group II: Is there a relationship to the development of intermediate-level glycopeptide resistance? J Infect Dis 2003; 187: 929-938.
- Jarraud S, Lyon GJ, Figueiredo JM, et al. Exofoliatin-producing isolates define a forth agr specificity group in S. aureus. J Bacteriol 2000; 182: 6517-6522.
- Koehl JL, Muthaiyan A, Jayaswal RK, et al. Cell wall composition and decreased autolytic activity and lysostaphin susceptibility of glycopeptide-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 2004; 48: 3749-3757.
- Moise-Broder PA, Sakoulas G, Eliopoulos GM, et al. Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin Infect Dis 2004; 38: 1700-1705.
- Pearman JW. Community-acquired MRSA: The Australian experience. In proceedings: The 10th international symposium on Staphylococci and Staphylococcal diseases. Tsukuba, Japan 2002; 18.
- Dufour P, Gillet Y, Bes M, et al. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: Emergence of a single clone that produces Panton-Valentine leukocidin. Clin Infect Dis 2002; 35: 819-824.
- Sakoulas G, Eliopoulos GM, Mollering RC, et al. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduces susceptibility to vancomycin. Antimicrob Agents Chemother 2002; 46: 1492-1502.
- Georke C, Kummel M, Dietz K, et al. Evaluation of interspecies interference due to agr polymorphism in Staphylococcus aureus during infection and colonization. J Infect Dis 2003; 188: 250-256.

