Document Type : Original Article
Authors
1 Infectious and Tropical Disease Research Center, Hormozgan Health Institute, Hormozgan University of Medical Sciences, Bandar Abbas, Iran
2 Research Center for Tropical and Infectious Diseases, Kerman University of Medical Sciences, Kerman, Iran
3 Department of Pathobiology, Faculty of Veterinary Medicine, Shahid Bahonar University, Kerman, Iran
4 Zoonosis Research Committee, Kerman University of Medical Sciences, Kerman, Iran
Abstract
Colibacillosis caused by avian pathogenic Escherichia coli (APEC) is an economic threat to the poultry industry throughout the world. Some of the virulence genes may enhance the ability of E. coli isolates to grow in the tissues of broilers. The APEC strains are assigned to a few distinct phylogenetic groups. The purpose of the present study was to detect the virulence genes and phylogenetic groups of E. coli isolates from colibacillosis cases in Japanese quail in 2014 in Kerman, Iran. In the present study, one hundred and two E. coli isolates were obtained from dead Japanese quails with colibacillosis. E. coli isolates were confirmed by standard biochemical and bacteriological methods. DNA of E. coli isolates was extracted by boiling method. The confirmed E. coli isolates were investigated to detect the phylogenetic groups and virulence genes including sfa/focDE, afaIBC, papEF by PCR methods. E. coli isolates were classified into A (62 isolates), B1 (24 isolates), B2 (12 isolates) and D (four isolates) phylogenetic groups. Among examined isolates nine isolates (8.82%) were positive for papE-F, five isolates (4.90%) for afaIB-C and two isolates (1.96%) for sfa/focD-E genes. Based on our findings, E. coli isolates from colibacillosis of Japanese quail could be assigned to various phylogenetic groups (mostly A and D), and they may contain the adhesion genes in a low prevalence.
Keywords
Main Subjects
Introduction
Avian pathogenic Escherichiacoli (APEC) are responsible for a variety of extra-intestinal pathogens in poultry, including colibacillosis, yolk sac infection, cellulitis, coli-granuloma and omphalitis.1 Although Japanese quail (Coturnix coturnix japonica) are reported as resistant birds against many diseases, APEC strains have been isolated from colisepticemic poultry with colibacillosis.1 Colibacillosis is an economic threat to the poultry industry which is a worldwide infection.2 Virulence factors (VFs) of extra-intestinal pathogenic E. coli (ExPEC) participate in colonization, cellular invasion and consequently reduction of the host immunity responses.3 Some of the VFs such as fimbrial antigens (P, AC/I, F1A and Stg), iron acquisition systems (aerobactin) and toxins (cytolethal distending toxins and hemolysins) may enhance the ability of E. coli to grow in the tissues of broilers.4 Expression of adhesions such as S and P fimbriae are considered to be an essential factor in pathogenesis of these strains because of their fundamental abilities for the adherence to the epithelium cells of birds.5 Stordeur et al. reported fimbrial and afimbrial adhesion genes normally expressed in extra-intestinal and intestinal strains isolated from birds.6 P fimbriae are an important step for the beginning and expansion of human urinary tract infections, but their role in pathogenesis of avian isolates has not been elucidated, completely.5 The E. coli strains are genetically diverse, and strains have been divided into four major phylogenetic groups (A, B1, B2, and D).7 According to the phylogenetic analysis most ExPEC strains are assigned to phylogenetic group B2 and to a lesser extent, to group D and possess a panel of VFs such as adhesins.8,9
The aims of present study were screening of the virulence genes papE-F, sfa/focD-E and afaIB-C and phylogenetic grouping of E. coli isolates from colibacillosis cases in Japanese quail in Kerman, Iran.
Materials and Methods
Bacterial isolates. In this cross-sectional study, a total number of 212 dead cases of Japanese quail were collected from the Eslam-Kish farm in Kerman province in 2014. Dead Japanese quail swabs were collected aseptically for the isolation of bacteria. Isolation of E coli was done from heart blood, liver, or typical visceral lesions in a fresh carcass. Samples were streaked on MacConkey and eosin-methylene blue (EMB) agar (Biolife Laboratories, Milan, Italy). Escherichia coli isolates were confirmed by standard biochemical and bacteriological methods. All of the E. coli isolates were stored in Luria-Bertani broth (Invitrogen, Paisley, Scotland) with 25% sterile glycerol (Pars Behbood Asia, Mashhad, Iran) at –70 ˚C.
Detection of virulence genes. DNA was extracted from the E. coli isolates and reference strains by the boiling method.The E. coli isolates were analyzed for the presence of the papE-F, sfa/focD-E and afaIB-C genes by PCR method as described previously.10 The reference strains were used as positive controls for virulence genes including 28C (papE-F), A30 (afaIB-C) and J96 (sfa/focD-E). The E. coli strain MG1655 was used as a negative control.
Phylogenetic groups. The phylogenetic analyses of the isolates were determined by presence and/or absence of the three genetic markers, chuA, yjaA, and TSPE4.C2 by a triplex PCR as described by Clermont et al.7 The isolates of E. coli were segregated in four distinct phylogenetic groups: A, B1, B2 and D. Four E. coli strains from the ECOR collection were used as controls for phylogenetic determination: ECOR58 (B1 group), ECOR62 (B2 group), ECOR50 (D group) and E. coli strain MG1655 as a positive control for phylogenetic ECOR group A. The primers used for amplification of the virulence genes and phylogenetic groups are shown in Table 1. The reference strains were from the bacterial culture collection, Department of Microbiology, School of Veterinary, Toulouse, France.
Table 1. Oligonucleotide primers used in this study.
|
Genes |
Primer Sequence (5′-3′) |
Product size (bp) |
|
afaIBC |
GCTGGGCAGCAAACTGATAACTCTC |
750 bp |
|
CATCAAGCTGTTTGTTCGTCCGCCG |
||
|
sfa/focDE |
CTCCGGAGAACTGGGTGCATCTTAC |
410 bp |
|
CGGAGGAGTAATTACAAACCTGGCA |
||
|
papEF |
GCAACAGCAACGCTGGTTGCATCAT |
336 bp |
|
AGAGAGAGCCACTCTTATACGGACA |
||
|
yjaA |
TGAAGTGTCAGGAGACGCTG |
211 bp |
|
ATGGAGAATGCGTTCCTCAAC |
||
|
TspE4C2 |
GAGTAATGTCGGGGCATTCA |
152 bp |
|
CGCGCCAACAAAGTATTACG |
||
|
chuA |
GACGAACCAACGGTCAGGAT |
279 bp |
|
TGCCGCCAGTACCAAAGACA |
Results
Among 212 dead cases of Japanese quail, pure colonies of E. coli were obtained from 102 samples in MacConkey and EMB agar. The recovered isolates were confirmed as E. coli based on standard bacteriological and biochemical tests. PCR analysis indicated that the 102 E. coli isolates assigned to the phylo-groups A (62 isolates; 60.78%), B1 (24 isolates; 23.52%), B2 (12 isolates; 11.76%) and D (4 isolates; 3.92%), (Fig.1).
Virulence genotyping of E. coli isolates showed that fourteen of the isolates exhibited at least one of the virulence genes. Multiplex PCR assay revealed that nine isolates (8.82%) were positive for papE-F, five isolates (4.90%) for afaIB-C and two isolates (1.96%) for sfa/focD-E genes (Figs. 2 and 3).
Overall, out of 102 E. coli isolates, nine isolates (8.82%) were positive for P fimbriae coding gene. These isolates segregated in phylogenetic groups A (two isolates; 22.22%), B1 (one isolate; 11.11%), B2 (5 isolates; 55.55%) and D (one isolate; 11.11%). afaIB-C gene was detected in 4.90% of isolates, which fell into A (40.00%), B1 (20.00%) and B2 (40.00%) phylogenetic groups. Two positive isolates for S fimbriae coding fell into B1 phylogenetic group. Two isolates were positive for both papE-F and sfa/focD-E genes, which belonged to two phylo-groups including B1 (n = 1) and B2 (n = 2).
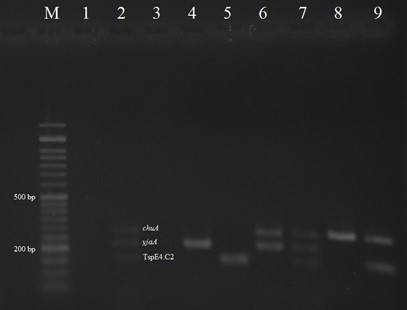
Fig. 1. Multiplex PCR results for the detection of E. coli phylogenetic groups among the colibacillosis cases of Japanese quail; Lane M: Ladder 50 bp; Lane 1: Negative control E. coli MG1655; Lane 2: Positive control E. coli ECOR62; Lanes 3, 4: A phylo-group; Lane 5: B1 phylo-group; Lanes 6, 7: B2 phylo-group; Lanes 8, 9: D phylo-group.

Fig. 2. The multiplex PCR results for sfa/focD-E and papE-F genes; Lane M: ladder 1Kb; Lane 1: Positive control E. coli 28C; Lane 2: Positive control E. coli J96; Lane 3: Negative control E. coli MG1655; Lanes 4, 5, 8: Positive isolates for sfa/focD-E gene; Lane 7: the Positive isolate for both sfa/focD-E and papE-F genes.
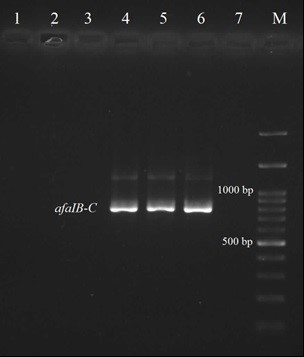
Fig. 3. The PCR results for afaIB-C gene; Lane M: Ladder 100 bp; Lanes 4, 5: Positive isolates for afaIB-C gene; Lane 6: Positive control E. coli A30; Lane 7: Negative control E. coli MG1655.
Discussion
APEC pathotype can cause localized and systemic infections in poultry, but the pathophysiology of these diseases has remained unknown. Fimbrial and putative colonization factors in APECs is considered to be an essential step in their pathogenicity and is associated with resistance to heterophil cells activity.5
In this study, 8.82% of isolates were positive for papE-F gene. Stordeur et al. reported pap sequence in 91.30% of avian E. coli isolates.6 The pap genes have also been observed in a considerable frequency in APEC strains in the present and other studies.11,12 Some studies suggested an important role for P fimbria in pathogenicity of APECs.5 In Ireland, 41.20% of isolates from septicemic birds were positive for pap genes, compared to 15.60% from E. coli isolated from healthy birds.13 Pourbakhsh et al. showed the in vivo expression of P fimbriae in experimentally inoculated chickens and suggested that P fimbriae may be involved in colonization and development of septicemia.14
In the present study, five and two isolates were positive for afaIB-C and sfa/focD-E genes, respectively. Results of the present study is comparable with other reports, S and afa fimbriae coding genes have been detected less than 10.00% of APEC isolates.4,15 The S fimbria has been seldom founded in APECs and its pathogenesis is not exactly clear in poultry colibacillosis.16 A study showed a low prevalence of afa (5.50%) and sfa (4.40%) genes in APECs and suggested that the factors may have a considerable role in colonization.6 In Iran, Salehi and Ghanbarpour have reported the presence of virulence genes in fecal E. coli isolates from colisepticemic cases of Japanese quail and found the genes afaI B-C, sfa/focD-E and papE-F in one, four, and 10 isolates, respectively, which is similar to the results of the current study.4
Genotyping of APEC and evolutionary study of the adhesive capacity of E. coli strains isolated from avian colibacillosisindicated that these isolates can be classified into various phylogenetic groups.4 In the present study, the E. coli isolates mostly fell into the phylo-groups A followed by B1. Ewers et al. reported a higher amount (46.10%) of E. coli isolates from APEC that was belonged to A followed by B2 (35.10%) phylo-group.15 Another study on phylogenetic groups in E. coli strains isolated from septicemic broiler and layer cases indicated that the isolates were belonged to A (71.00%), B1 (4.10%), B2 (7.90%) and D (18.70%) phylo-groups.17 The phylogenetic background of 109 E. coli isolates from heart blood samples of dead quail has been evaluated by Salehi and Ghanbarpour in Iran in which 55.00% of isolates were belonged to A and the remaining were B1 (18.30%), B2 (17.40%) and D (9.20%) phylo-groups, that was similar to our results.4
According to the results of the present study, E. coli isolates from colibacillosis of Japanese quail were distributed in different phylogenetic groups, which contained few adhesins genes. However, avian colibacillosis might be associated with other virulence genes that was not examined. Further studies are needed to survey the phylogenetic background of E. coli isolates in comparison with their virulence genes to further refine the definition of pathogenic E. coli. Regarding limitation in collection of data in this species, findings of the present study could be helpful to understand the prevention and control of APEC in quail and to develop new and improved vaccines.
Acknowledgments
The authors are thankful to Dr. Eric Oswald from School of Veterinary, Toulouse, France, for providing the reference strains.
- Ghanbarpour R, Salehi M, Oswald E. Virulence genotyping of Escherichia coli isolates from avian cellulitis in relation to phylogeny. Comp Clin Path 2010; 19(2): 147-153.
- Roy P, Purushothaman V, Koteeswaran A, et al. Isolation, characterization, and antimicrobial drug resistance pattern of Escherichia coli isolated from Japanese quail and their environment. J Appl Poultry Res 2006; 15(3): 442-446.
- Adib N, Ghanbarpour R, Solatzadeh H, et al. Antibiotic resistance profile and virulence genes of uropathogenic Escherichia coli isolates in relation to phylogeny. Trop Biomed 2014; 31(1): 17-25.
- Salehi M, Ghanbarpour R. Phenotypic and genotypic properties of Escherichia coli isolated from colisepticemic cases of Japanese quail. Trop Anim Health Prod 2010; 42(7): 1497-1504.
- de Campos TA, Stehling EG, Ferreira A, et al. Adhesion properties, fimbrial expression and PCR detection of adhesin-related genes of avian Escherichia coli strains. Vet Microbiol 2005; 106(3): 275-285.
- Stordeur P, Marlier D, Blanco J, et al. Examination of Escherichia coli from poultry for selected adhesin genes important in disease caused by mammalian pathogenic E. coli. Vet Microbiol 2002; 84(3): 231-241.
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 2000; 66(10): 4555-4558.
- Escobar-Páramo P, Clermont O, Blanc-Potard AB, et al. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol Biol Evol 2004; 21(6): 1085-1094.
- Bingen-Bidois M, Clermont O, Bonacorsi S, et al. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect Immun 2002; 70(6): 3216-3226.
- Yamamoto S, Terai A, Yuri K, et al. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol 1995; 12(2): 85-90.
- Knöbl T, Baccaro MR, Moreno AM, et al. Virulence properties of Escherichia coli isolated from ostriches with respiratory disease. Vet microbiol 2001; 83(1): 71-80.
- Ngeleka M, Brereton L, Brown G, et al. Pathotypes of avian Escherichia coli as related to tsh-, pap-, pil-, and iuc-DNA sequences, and antibiotic sensitivity of isolates from internal tissues and the cloacae of broilers. Avian Dis 2002; 46(1): 143-152.
- McPeake SJ, Smyth JA, Ball HJ. Characterisation of avian pathogenic Escherichia coli (APEC) associated with colisepticemia compared to fecal isolates from healthy birds. Vet Microbiol 2005; 110(3): 245-253.
- Pourbakhsh SA, Dho-Moulin M, Brée A, et al. Localization of thein vivoexpression of P and F1 fimbriae in chickens experimentally inoculated with pathogenic Escherichia coli. Microb Pathog 1997; 22(6): 331-341.
- Ewers C, Li G, Wilking H, et al. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: How closely related are they? Int J Med Microbiol 2007; 297(3): 163-176.
- Knöbl T, Gomes TA, Vieira MA, et al. Occurrence of adhesin-encoding operons in Escherichia coli isolated from breeders with salpingitis and chicks with omphalitis. Braz J Microbiol 2006; 37(2): 140-143.
- Dissanayake DR, Wijewardana TG, Gunawardena GA, et al. Distribution of lipopolysaccharide core types among avian pathogenic Escherichia coli in relation to the major phylogenetic groups. Vet Microbiol 2008; 132(3): 355-363.

